
The results obtained are consistent with other genomic studies and can be corroborated with the involved pathways and biological functions.

For further evaluation, we used the Glioblastoma multiforme (GBM) dataset and identified the differentially expressed genes in each of the subtypes. Furthermore, we compared the effect of feature selection and similarity measures for subtype detection. We also predicted the optimal number of subtypes in a cancer type using the silhouette score and found that the detected subtypes exhibit significant differences in survival profiles. We performed cancer subtype detection on four different cancer types from The Cancer Genome Atlas (TCGA) datasets using four autoencoder implementations. In this paper, we compared the performance of different deep learning autoencoders for cancer subtype detection. Different data fusion and subtyping approaches have been proposed over the years, such as kernel-based fusion, matrix factorization, and deep learning autoencoders.
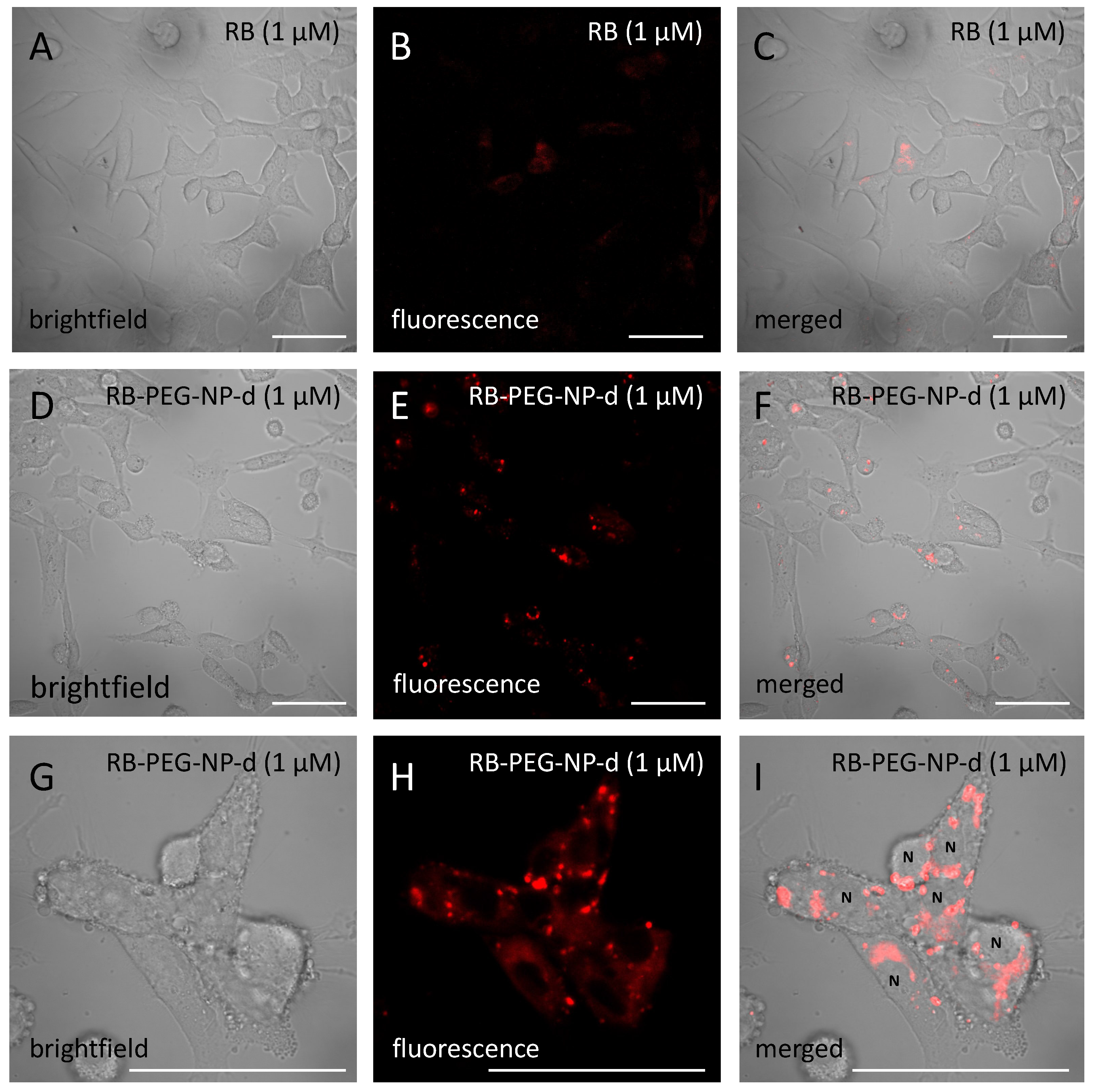
Subtype detection is often a complex problem, and in most cases, needs multi-omics data fusion to achieve accurate subtyping. Therefore, cancer subtype detection using genomics level data is a significant research problem.

Depending upon the activated pathway(s), the survival of the patients varies significantly and shows different efficacy to various drugs. A heterogeneous disease such as cancer is activated through multiple pathways and different perturbations.


 0 kommentar(er)
0 kommentar(er)
